1Division of Pulmonary and Vital Care Medication, Zhengzhou College Individuals’s Hospital, Henan Provincial Individuals’s Hospital, Zhengzhou, Henan, 450003, Individuals’s Republic of China; 2Division of Pulmonary and Vital Care Medication, Central China Fuwai Hospital, Central China Fuwai Hospital of Zhengzhou College, Zhengzhou, Henan, 451464, Individuals’s Republic of China; 3Division of Well being Administration, Henan Provincial Individuals’s Hospital, Zhengzhou College Individuals’s Hospital, Zhengzhou, Henan, 450003, Individuals’s Republic of China; 4Division of Pulmonary and Vital Care Medication, Zhengzhou College Individuals’s Hospital, Henan Provincial Individuals’s Hospital, Academy of Medical Science, Zhengzhou College, Zhengzhou, Henan, 450003, Individuals’s Republic of China; 5Division of Pulmonary and Vital Care Medication, Henan College Individuals’s Hospital, Henan Provincial Individuals’s Hospital, Zhengzhou, Henan, 450003, Individuals’s Republic of China; 6Division of Pulmonary and Vital Care Medication, Zhengzhou College Individuals’s Hospital, Henan Provincial Individuals’s Hospital, Henan College Individuals’s Hospital, Zhengzhou, Henan, 450003, Individuals’s Republic of China
Correspondence: Yong Qi, Division of Pulmonary and Vital Care Medication, Zhengzhou College Individuals’s Hospital, Henan Provincial Individuals’s Hospital, Henan College Individuals’s Hospital, No. 7 Weiwu Street, Jinshui District, Zhengzhou, 450003, Individuals’s Republic of China, Tel +8615890110258, Electronic mail [email protected]
Background: Though the potential of coronavirus illness 2019 (COVID-19) sufferers to develop pulmonary embolism (PE) is well known, the underlying mechanism has not been utterly elucidated. This research aimed to establish genes frequent to COVID-19 and PE to disclose the underlying pathogenesis of susceptibility to PE in COVID-19 sufferers.
Strategies: COVID-19 genes had been obtained from the GEO database and the OMIM, CTD, GeneCards, and DisGeNET databases; PE genes had been obtained from the OMIM, CTD, GeneCards, and DisGeNET databases. We overlapped the genes of COVID-19 and PE to acquire frequent genes for extra evaluation, together with practical enrichment, protein–protein interplay, and immune infiltration evaluation. Hub genes had been recognized utilizing cytoHubba, a plugin of Cytoscape, and validated utilizing the unbiased datasets GSE167000 and GSE13535. The genes validated by the above datasets had been additional validated in medical samples.
Outcomes: We obtained 36 genes shared by PE and COVID-19. Purposeful enrichment and immune infiltration analyses revealed the involvement of cytokines and immune activation. 5 genes (CCL2, CXCL10, ALB, EGF, and MKI67) had been recognized as hub genes frequent to COVID-19 and PE. CXCL10 was validated in each unbiased datasets (GSE167000 and GSE13535). Serum ranges of CXCL10 within the COVID-19 group and the COVID-19 mixed with PE group had been considerably increased than these within the wholesome management group (PConclusion: Our research reveals frequent genes shared by PE and COVID-19 and identifies CXCL10 as a potential reason behind susceptibility to PE in COVID-19 sufferers.
Introduction
Coronavirus illness 2019 (COVID-19), attributable to extreme acute respiratory syndrome coronavirus 2 (SARS-CoV-2) an infection, stays an acute infectious illness of worldwide pandemic proportions and is a public well being illness that critically endangers human well being.1,2 COVID-19 sufferers have a number of problems, together with pulmonary embolism (PE).3 PE refers back to the medical and pathophysiological syndrome wherein a thrombus happens in the primary trunk of the pulmonary artery or its branches, inflicting blockage and pulmonary circulation problems.4,5 PE, with excessive incidence, missed analysis and mortality charges, is the third commonest reason behind cardiovascular loss of life worldwide after stroke and coronary heart assault.6
Accumulating proof means that inpatients with COVID-19 extra incessantly develop PE occasions than inpatients with out COVID-19.7–9 A meta-analysis together with 3342 COVID-19 sufferers confirmed that the incidence of PE occasions in COVID-19 sufferers was 16.5%, significantly increased than that in sufferers with out COVID-19.10 After a meta-analysis of 13 postmortem research of COVID-19 sufferers, Zuin et al concluded that the potential of acute PE occasions in COVID-19 sufferers has been underestimated in medical apply.11 Not too long ago, a big Swedish cohort research revealed that in contrast with the management interval, the danger ratio for a primary PE throughout days 1–30 after COVID-19 was 33.05, and the elevated threat lasted for six months after COVID-19.12 Early within the COVID-19 epidemic, German researchers carried out autopsies on 12 sufferers who died from the illness and located PE to be the direct reason behind loss of life in 4 of them (1/3).13 Moreover, in contrast with the PE sufferers with out COVID-19 an infection, PE sufferers with COVID-19 had an elevated threat of in-hospital mortality, septic shock, respiratory failure, and an extended hospital keep.14
PE is broadly believed to be one of many types of venous thrombosis. Nonetheless, the concept PE is the results of the migration of blood clots from the venous system is questioned as a result of no preliminary thrombus is discovered in lots of sufferers.15–17 Histologic evaluation of pulmonary vessels in sufferers with COVID-19 reveals widespread thrombosis with microangiopathy,18 and in situ immunothrombosis performs a job within the pathophysiology of COVID-19-associated PE.19,20 This angle is supported by Katsoularis’ research wherein threat ratios throughout days 1–30 after COVID-19 had been 33.05, considerably exceeding that for deep-vein thrombosis (4.98).12 In a single research, conventional threat elements for venous thromboembolic illness weren’t related to the incidence of PE in COVID-19 sufferers.21 As well as, prophylactic anticoagulation didn’t forestall the incidence of PE in hospitalized sufferers.22 Due to this fact, it’s essential to particularly research COVID-19-associated PE quite than deal with it as a type of venous thrombosis.
At present, analysis on COVID-19-associated PE is dominated by observational research of incidence, prognosis, and imaging traits, whereas few research have been performed to research the mechanisms of COVID-19-associated PE. Bioinformatics evaluation supplies a way to discover the genetic relationship between two ailments. Earlier research have indicated that SARS-CoV-2 an infection could cause a myriad of problems as a result of there are frequent genes between COVID-19 and these problems.23–26 Due to this fact, this research aimed to discover the potential genetic relationship between COVID-19 and PE by bioinformatics to disclose the underlying pathogenesis of susceptibility to PE occasions in COVID-19 sufferers.
On this research, we used COVID-19 transcriptome knowledge from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database and COVID-19-related genes and PE-related genes from disease-related databases to carry out bioinformatics analyses after which validated frequent key genes between COVID-19 and PE. That is the primary genomics research of COVID-19 and PE.
Supplies and Strategies
Assortment of COVID-19 and PE Genes
By looking the OMIM (http://omim.org/), CTD (http://ctdbase.org/), GeneCards (https://www.genecards.org/), and DisGeNET (https://www.disgenet.org/) databases, we collected genes associated to COVID-19 and PE. As well as, microarray expression knowledge of COVID-19 sufferers had been obtained from the GEO database. GSE157103 was chosen, which consisted of 126 samples, together with 100 sufferers with COVID-19 and 26 sufferers with out COVID-19, consisting of each ICU and non-ICU sufferers. Sadly, as no human knowledge can be found within the GEO database, we solely obtained PE genes by the 4 disease-related databases talked about above.
We collected the highest 500 genes in every database based on every database scoring rule. If there have been fewer than 500 genes within the database, all of them had been included. The GSE157103 dataset was analyzed utilizing the DESeq2 package deal, and an adjusted p-value (false discovery charge) <0.05 and∣log2FoldChange∣>1 had been used because the screening standards for essential differentially expressed genes (DEGs).
Subsequently, we obtained COVID-19 genes by taking the intersection of the COVID-19-related genes within the disease-related database and the DEGs of GSE157103. We then overlapped the genes of COVID-19 and PE to acquire frequent genes for additional evaluation. We accessed these web sites on 4 March 2022.
Gene Enrichment Evaluation
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses had been used to research organic actions and pathways. To know the practical traits of the frequent genes of COVID-19 and PE, we executed GO and KEGG enrichment analyses utilizing R’s cluster profile package deal, with a p-value <0.05 because the cutoff.
Protein–Protein Interplay (PPI) Community Evaluation and Screening for Frequent Hub Genes
We constructed a PPI community based mostly on the STRING database (https://string-db.org/), with a medium confidence rating of > 0.4, and the PPI community was visualized by way of Cytoscape (Model 3.9.1).27 Moreover, we recognized hub genes by 5 algorithms (BottleNeck, Closeness, Radiality, Betweenness, Stress) of the cytoHubba plugin of Cytoscape. Frequent hub genes of COVID-19 and PE had been obtained by taking the intersection of the highest 10 genes in every of the 5 algorithms.
Immune Infiltration Evaluation and Correlations Between Hub Genes and Immune Cell Infiltration
Immune infiltration evaluation was carried out by way of the CIBERSORTx instrument,28 a deconvolution algorithm that evaluates the expression of associated genes based mostly on gene expression, to calculate the ratio of COVID-19 samples. Correlations between every immune cell and hub genes had been calculated utilizing the Sangerbox on-line instrument.29
Validation of Hub Genes in Exterior Datasets
To scale back the potential of false positives, we validated hub genes individually utilizing different COVID-19 and PE datasets from the GEO database. For COVID-19, we chosen GSE167000 for validation, which incorporates 65 SARS-CoV-2-positive sufferers and 30 SARS-CoV-2-negative sufferers. Moreover, we used the rat PE mannequin dataset GSE13535 for PE. GSE13535 consists of 6 management rats and 16 PE mannequin rats. Fashions with delicate PE had been established at 2 hours after modeling and fashions with extreme PE had been established at 18 hours later. Expression knowledge for the hub genes had been obtained utilizing the GEOquery package deal.
Verification of Genes by Medical Samples
To additional confirm the genes validated above, we collected 6 blood samples from COVID-19 mixed with PE inpatients. COVID-19 was decided by a constructive polymerase chain response check for SARS-CoV-2 and PE was recognized by computed tomography pulmonary angiography. As well as, we collected 6 blood samples from COVID-19-alone inpatients and 4 wholesome people who had been damaging for anti-SARS-CoV-2 IgM/IgG. The COVID-19-alone sufferers and the wholesome controls had been matched with the COVID-19 mixed with PE sufferers for age and intercourse. Blood samples had been obtained from Henan Provincial Individuals’s Hospital (Henan, China) and centrifuged at 1000×g for 10 min. Ranges of CXCL10 in serum had been measured utilizing a human ELISA equipment (Elabscience, Wuhan, China). This research was authorized by the ethics committee of Henan Provincial Individuals’s Hospital (Moral Assessment 2023(97)), and written knowledgeable consent from all members was offered for using their blood samples.
Statistical Evaluation
GraphPad Prism model 8.0.2 was used to research the information. The outcomes are displayed because the imply ± SD. Variations between the 2 teams had been in contrast by an unpaired Scholar′s t-test. A two-tailed p-value < 0.05 was thought-about statistically important.
Outcomes
Assortment of COVID-19 and PE Genes
We obtained 880 DEGs from GSE157103 and 500, 500, 500 and three COVID-19-related genes from the GeneCards, DisGeNET, CTD and OMIM databases, respectively. Seventy-two COVID-19 genes had been obtained by merging and deduplicating the outcomes collected from 4 databases and taking the intersection with DEGs from GSE157103. Equally, after merging and deduplicating 500, 93, 500, and 207 PE-related genes from the GeneCards, DisGeNET, CTD, and OMIM databases, respectively, we obtained 1069 PE genes. By taking the intersection of 72 COVID-19 genes with 1069 PE genes, we obtained 36 genes frequent to each COVID-19 and PE. The screening process and outcomes are proven in Desk 1 and Determine 1.
|
Desk 1 Assortment of COVID-19 and PE Genes |
Gene Enrichment Evaluation
The highest 10 GO phrases and KEGG pathways are summarized in Determine 2. GO annotation consists of three phrases: organic course of, cell element, and molecular operate. For organic processes, the frequent genes had been enriched in response to chemical and cytokine-mediated signaling pathways (Determine 2A). Cell element gadgets had been primarily enriched within the endomembrane system and protein-containing complicated (Determine 2B). The molecular operate class was primarily enriched in anion binding and carbohydrate by-product binding (Determine 2C). KEGG pathway enrichment evaluation included focal adhesion, the cell cycle, and the p53 signaling pathway (Determine 2D).
PPI Community Evaluation and Screening of Frequent Hub Genes
To discover protein interactions, we constructed a PPI community of 36 frequent genes utilizing the STRING database. There have been a complete of 36 nodes and 152 edges within the community, which was visualized by Cytoscape (Determine 3A). We screened the highest 10 hub genes with 5 algorithms from the cytoHubba plugin. 5 hub genes (CCL2, CXCL10, ALB, EGF, and MKI67) had been obtained by taking the intersection of the highest 10 genes within the 5 algorithms (Determine 3B). CCL2, CXCL10, EGF, and MKI67 had been upregulated in GSE157103; ALB, encoding essentially the most considerable protein in human blood, was downregulated (Desk 2).
 |
Desk 2 Hub Genes |
Immune Infiltration Evaluation and Correlations Between Hub Genes and Immune Cells
There may be proof that immune cell infiltration is concerned within the growth of COVID-19.30 The CIBERSORTx algorithm has been utilized to research the panorama of immune infiltration in COVID-19. The proportions of twenty-two immune cell sorts in COVID-19 are proven in Determine 4A. Particularly, the forms of immune cells in COVID-19 had been B cells, plasma cells, naive CD4+ T cells, activated reminiscence CD4+ T cells, regulatory T cells, monocytes, and resting mast cells. As well as, we analyzed the correlation between every immune cell and the 5 hub genes. As proven in Determine 4B, CCL2 expression was related to resting reminiscence CD4+ T cells, gamma delta T cells, and neutrophils; CXCL10 correlated with activated reminiscence CD4+ T cells and activated dendritic cells; ALB correlated with naive CD4+ T cells; EGF correlated with activated reminiscence CD4+ T cells and Tregs; and MKI67 correlated with plasma cells and activated reminiscence CD4+ T cells. In abstract, these outcomes recommend that CCL2, CXCL10, ALB, EGF, and MKI67 might contribute to the immune microenvironment of COVID-19.
Validation of Hub Genes in Exterior Datasets
To validate the reliability of the 5 hub genes, we first verified the expression of those genes within the GSE167000 dataset. As proven in Determine 5A, CXCL10, EGF, and MKI67 on this dataset had been considerably upregulated within the SARS-CoV-2-positive group in contrast with the SARS-CoV-2- damaging group. Within the GSE13535 dataset, the expression ranges of CCL2 and CXCL10 had been considerably elevated within the PE mannequin group in contrast with the management group, each at 2 and 18 hours after modeling. The outcomes are proven in Determine 5B and C. In abstract, solely CXCL10 was discrepant in each exterior datasets. Due to this fact, we take into account CXCL10 to be the almost certainly key gene concerned in COVID-19 and PE.
Verification of Serum CXCL10 Ranges in Human Samples
To confirm whether or not CXCL10 is a key gene concerned in COVID-19 related to PE, we validated CXCL10 with blood samples. The protein encoded by CXCL10 is a secreted protein, often known as interferon-inducible protein 10 (IP-10), which belongs to the CXC chemokine household of chemokines and is current in serum and plasma. We measured serum ranges of CXCL10 in 4 wholesome controls, 6 COVID-19 sufferers, and 6 COVID-19 sufferers with PE. Ranges of CXCL10 within the COVID-19 and the COVID-19 mixed with PE teams had been considerably increased than these within the wholesome management group (p <0.001). As well as, there have been important variations between the COVID-19 group and the COVID-19 mixed with PE group (p <0.01), as proven in Determine 6.
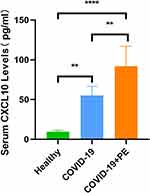 |
Determine 6 Verification of serum CXCL10 ranges in human samples (4 wholesome controls, 6 COVID-19 sufferers, and 6 COVID-19 sufferers with PE). **p < 0.01, ****p < 0.001. |
Dialogue
Throughout the COVID-19 pandemic, a number of research have described an elevated threat of growing PE, which contributes to a big improve in mortality amongst COVID-19 sufferers. Nonetheless, the underlying pathogenesis of susceptibility to PE in COVID-19 just isn’t but totally understood. On this research, we first carried out a genomics evaluation of COVID-19 and PE to disclose genetic interrelations between the 2 ailments.
We recognized 36 frequent genes shared in COVID-19 and PE. In a way, these frequent genes recommend that there are relationships between COVID-19 and PE. Enrichment evaluation confirmed that these genes had been concerned in cytokine-mediated signaling pathways, and immune infiltration evaluation indicated that immune activation was concerned in COVID-19. SARS-CoV-2 can repeatedly activate the physique’s pure immune system and, consequently, trigger the discharge of inflammatory cytokines.30,31 Cytokines play important roles within the pathogenesis of COVID-19.32–34 Particularly, uncontrolled cytokines might result in a systemic inflammatory response syndrome and launch “messenger substances”, which trigger thrombosis and blood vessel blockage.13 That is in step with medical research wherein histologic evaluation of the lung in sufferers with COVID-19 confirmed infiltration of a number of immune cells and widespread microthrombosis.18,35,36 Irritation could also be a serious contributor to PE within the context of COVID-19.
We then carried out a complete evaluation to assemble the PPI community, and a powerful genetic, protein-based relationship was discovered amongst frequent genes shared in COVID-19 and PE. CCL2, CXCL10, ALB, EGF, and MKI67 had been recognized as hub genes, with CXCL10 validated by exterior datasets and human samples. These outcomes recommend that CXCL10 is a key gene frequent to each COVID-19 and PE and that it could be chargeable for susceptibility to PE in COVID-19 sufferers.
CXCL10, additionally termed IP-10, is secreted after interferon-gamma manufacturing by all kinds of cell sorts, corresponding to endothelial cells, fibroblasts, monocytes, and T lymphocytes.37 CXCL10 is a key regulator of the “cytokine storms” attributable to SARS-CoV-2 an infection38 and tends to be elevated earlier in COVID-19 sufferers than different inflammatory cytokines.39 There may be ample proof that in COVID-19 sufferers, CXCL10 ranges are considerably elevated and related to hostile medical outcomes.40–42 According to these findings, our research confirmed elevated ranges of CXCL10 within the COVID-19 group with or with out PE in contrast with wholesome controls.
The function of CXCL10 in atherosclerosis and myocardial infarction has been extensively described.43,44 Nonetheless, it isn’t clear whether or not CXCL10 performs a job within the growth of PE. Research have proven that CXCL10 induces not solely chemoattraction of inflammatory cells but in addition migration and proliferation of endothelial cells and vascular easy muscle cells.45,46 CXCR3, the receptor for CXCL10, has been reported to be expressed in pulmonary artery endothelial cells (PAECs), and CXCL10 can result in PAEC dysfunction.47 Improved endothelial therapeutic prevents arterial thrombosis, however CXCL10 can inhibit endothelial therapeutic.48 Due to this fact, CXCL10 could also be concerned within the formation of arterial thrombosis. In our research, the CXCL10 ranges within the COVID-19 mixed with PE group had been considerably increased than these within the COVID-19 group.
In abstract, we hypothesize that CXCL10, as an early upregulated inflammatory consider COVID-19, performs a twin function by recruiting numerous inflammatory cells to infiltrate and launch “messenger substances”, selling thrombosis, whereas additionally doubtlessly inflicting dysfunction of PAECs and contributing to pulmonary thrombosis in COVID-19 sufferers. Sooner or later, we are going to deal with elucidating the underlying mechanisms by which CXCL10 might contribute to the event of pulmonary artery thrombosis in COVID-19 sufferers.
Limitations of the Examine
The research has a number of limitations, which should be acknowledged. Given the paucity of accessible human datasets for PE, we used the dataset from rats to validate hub genes. Nonetheless, CXCL10 has additionally been validated in medical samples. As well as, restricted medical samples had been accessible for this research, and we solely in contrast CXCL10 ranges in 4 wholesome controls, 6 COVID-19 sufferers, and 6 COVID-19 sufferers with PE. We’ll additional validate our conclusions by increasing the pattern dimension.
Conclusion
On this research, we obtained 36 frequent genes, established a coexpression community between COVID-19 and PE, and recognized and validated CXCL10 as a typical key gene in each ailments which may be chargeable for susceptibility to PE occasions in COVID-19 sufferers. Consequently, our analysis first describes the potential genetic relationship between COVID-19 and PE to additional reveal the mechanisms of COVID-19-associated PE.
Abbreviations
COVID-19, coronavirus illness 2019; SARS-CoV-2, extreme acute respiratory syndrome coronavirus 2; PE, pulmonary embolism; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interplay; PAECs, pulmonary artery endothelial cells.
Knowledge Sharing Assertion
We offer particulars of the supplies and strategies in our manuscript.
Moral Approval and Consent to Take part
This research was carried out in accordance with the Helsinki declaration and was authorized by the ethics committee of Henan Provincial Individuals’s Hospital (Moral Assessment 2023(97)), and written knowledgeable consent from all members was offered for using their blood samples.
Acknowledgments
We sincerely thank the general public databases GEO, OMIM, CTD, GeneCards, DisGeNET, and STRING for offering knowledge for our analysis and Cytoscape, CIBERSORTx, and Sangerbox for analyzing knowledge.
Funding
This research is supported by the Pure Science Basis of Henan Province (Yong Qi, No. 232300421122).
Disclosure
The authors declare that they haven’t any conflicts of curiosity.
References
1. Yuan Y, Jiao B, Qu L, et al. The event of COVID-19 therapy. Entrance Immunol. 2023;14:1125246. doi:10.3389/fimmu.2023.1125246
2. Zhang JJ, Dong X, Liu GH, et al. Threat and protecting elements for COVID-19 morbidity, severity, and mortality. Clin Rev Allergy Immunol. 2023;64(1):90–107. doi:10.1007/s12016-022-08921-5
3. Dou Q, Wei X, Zhou Ok, et al. Cardiovascular manifestations and mechanisms in sufferers with COVID-19. Traits Endocrinol Metab. 2020;31(12):893–904. doi:10.1016/j.tem.2020.10.001
4. Agnelli G, Becattini C. Anticoagulant therapy for acute pulmonary embolism: a pathophysiology-based medical strategy. Eur Respir J. 2015;45(4):1142–1149. doi:10.1183/09031936.00164714
5. Essien EO, Rali P, Mathai SC. Pulmonary Embolism. Med Clin North Am. 2019;103(3):549–564. doi:10.1016/j.mcna.2018.12.013
6. Konstantinides SV, Meyer G, Becattini C, et al. 2019 ESC Tips for the analysis and administration of acute pulmonary embolism developed in collaboration with the European Respiratory Society (ERS). Eur Coronary heart J. 2020;41(4):543–603. doi:10.1093/eurheartj/ehz405
7. Poissy J, Goutay J, Caplan M, et al. Pulmonary embolism in sufferers with COVID-19: consciousness of an elevated prevalence. Circulation. 2020;142(2):184–186. doi:10.1161/CIRCULATIONAHA.120.047430
8. Helms J, Tacquard C, Severac F, et al. Excessive threat of thrombosis in sufferers with extreme SARS-CoV-2 an infection: a multicenter potential cohort research. Intensive Care Med. 2020;46(6):1089–1098. doi:10.1007/s00134-020-06062-x
9. Piroth L, Cottenet J, Mariet AS, et al. Comparability of the traits, morbidity, and mortality of COVID-19 and seasonal influenza: a nationwide, population-based retrospective cohort research. Lancet Respir Med. 2021;9(3):251–259. doi:10.1016/S2213-2600(20)30527-0
10. Suh YJ, Hong H, Ohana M, et al. Pulmonary embolism and deep vein thrombosis in COVID-19: a scientific evaluate and meta-analysis. Radiology. 2021;298(2):E70–E80. doi:10.1148/radiol.2020203557
11. Zuin M, Engelen MM, Bilato C, et al. Prevalence of acute pulmonary embolism at post-mortem in sufferers with COVID-19. Am J Cardiol. 2022;171:159–164. doi:10.1016/j.amjcard.2022.01.051
12. Katsoularis I, Fonseca-Rodriguez O, Farrington P, et al. Dangers of deep vein thrombosis, pulmonary embolism, and bleeding after covid-19: nationwide self-controlled circumstances sequence and matched cohort research. BMJ. 2022;377:e069590. doi:10.1136/bmj-2021-069590
13. Wichmann D, Sperhake JP, Lutgehetmann M, et al. Post-mortem findings and venous thromboembolism in sufferers with COVID-19: a Potential Cohort Examine. Ann Intern Med. 2020;173(4):268–277. doi:10.7326/M20-2003
14. Safiriyu I, Fatuyi M, Mehta A, et al. Impression of COVID-19 an infection on the medical outcomes of pulmonary embolism hospitalizations: a nationwide evaluation. Curr Probl Cardiol. 2023;48(7):101669. doi:10.1016/j.cpcardiol.2023.101669
15. Tadlock MD, Chouliaras Ok, Kennedy M, et al. The origin of deadly pulmonary emboli: a postmortem evaluation of 500 deaths from pulmonary embolism in trauma, surgical, and medical sufferers. Am J Surg. 2015;209(6):959–968. doi:10.1016/j.amjsurg.2014.09.027
16. Ahuja J, Shroff GS, Benveniste MF, et al. In situ pulmonary artery thrombosis: unrecognized complication of radiation remedy. AJR Am J Roentgenol. 2020;215(6):1329–1334. doi:10.2214/AJR.19.22741
17. Cao Y, Geng C, Li Y, et al. In situ pulmonary artery thrombosis: a beforehand ignored illness. Entrance Pharmacol. 2021;12:671589. doi:10.3389/fphar.2021.671589
18. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi:10.1056/NEJMoa2015432
19. Poor HD. Pulmonary thrombosis and thromboembolism in COVID-19. Chest. 2021;160(4):1471–1480. doi:10.1016/j.chest.2021.06.016
20. Quartuccio L, Sonaglia A, Casarotto L, et al. Medical, laboratory and immunohistochemical characterization of in situ pulmonary arterial thrombosis in deadly COVID-19. Thromb Res. 2022;219:95–101. doi:10.1016/j.thromres.2022.09.012
21. Fauvel C, Weizman O, Trimaille A, et al. Pulmonary embolism in COVID-19 sufferers: a French multicentre cohort research. Eur Coronary heart J. 2020;41(32):3058–3068. doi:10.1093/eurheartj/ehaa500
22. Bompard F, Monnier H, Saab I, et al. Pulmonary embolism in sufferers with COVID-19 pneumonia. Eur Respir J. 2020;56(1). doi:10.1183/13993003.01365-2020
23. Chen Z, Chen C, Chen F, et al. Bioinformatics evaluation of potential pathogenesis and threat genes of immunoinflammation-promoted renal harm in extreme COVID-19. Entrance Immunol. 2022;13:950076. doi:10.3389/fimmu.2022.950076
24. Li Y, Liu Y, Duo M, et al. Bioinformatic evaluation and preliminary validation of potential therapeutic targets for COVID-19 an infection in bronchial asthma sufferers. Cell Commun Sign. 2022;20(1):201. doi:10.1186/s12964-022-01010-2
25. Lv Y, Zhang T, Cai J, et al. Bioinformatics and techniques biology strategy to establish the pathogenetic hyperlink of Lengthy COVID and Myalgic Encephalomyelitis/Persistent Fatigue Syndrome. Entrance Immunol. 2022;13:952987. doi:10.3389/fimmu.2022.952987
26. Zhang W, Di L, Liu Z, et al. TIMELESS is a key gene mediating thrombogenesis in COVID-19 and antiphospholipid syndrome. Sci Rep. 2022;12(1):17248. doi:10.1038/s41598-022-21694-3
27. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software program surroundings for built-in fashions of biomolecular interplay networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
28. Chen B, Khodadoust MS, Liu CL, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Strategies Mol Biol. 2018;1711:243–259. doi:10.1007/978-1-4939-7493-1_12
29. Shen W, Track Z, Zhong X, et al. Sangerbox: a complete, interaction-friendly medical bioinformatics evaluation platform. iMeta. 2022;1(3):e36. doi:10.1002/imt2.36
30. Mehta P, McAuley DF, Brown M, et al. COVID-19: take into account cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi:10.1016/S0140-6736(20)30628-0
31. Ramasamy S, Subbian S. Vital determinants of cytokine storm and Sort I interferon response in COVID-19 pathogenesis. Clin Microbiol Rev. 2021;34(3). doi:10.1128/CMR.00299-20
32. Coperchini F, Chiovato L, Ricci G, et al. The cytokine storm in COVID-19: additional advances in our understanding the function of particular chemokines concerned. Cytokine Progress Issue Rev. 2021;58:82–91. doi:10.1016/j.cytogfr.2020.12.005
33. Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, et al. SARS-CoV-2 an infection: the function of cytokines in COVID-19 illness. Cytokine Progress Issue Rev. 2020;54:62–75. doi:10.1016/j.cytogfr.2020.06.001
34. Hsu RJ, Yu WC, Peng GR, et al. The function of cytokines and chemokines in extreme acute respiratory syndrome Coronavirus 2 infections. Entrance Immunol. 2022;13:832394. doi:10.3389/fimmu.2022.832394
35. Wang S, Yao X, Ma S, et al. A single-cell transcriptomic panorama of the lungs of sufferers with COVID-19. Nat Cell Biol. 2021;23(12):1314–1328. doi:10.1038/s41556-021-00796-6
36. Khismatullin RR, Ponomareva AA, Nagaswami C, et al. Pathology of lung-specific thrombosis and irritation in COVID-19. J Thromb Haemost. 2021;19(12):3062–3072. doi:10.1111/jth.15532
37. Luster AD, Ravetch JV. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10). J Exp Med. 1987;166(4):1084–1097. doi:10.1084/jem.166.4.1084
38. Zhang N, Zhao YD, Wang XM. CXCL10 an essential chemokine related to cytokine storm in COVID-19 contaminated sufferers. Eur Rev Med Pharmacol Sci. 2020;24(13):7497–7505. doi:10.26355/eurrev_202007_21922
39. Lu Q, Zhu Z, Tan C, et al. Modifications of serum IL-10, IL-1beta, IL-6, MCP-1, TNF-alpha, IP-10 and IL-4 in COVID-19 sufferers. Int J Clin Pract. 2021;75(9):e14462. doi:10.1111/ijcp.14462
40. Gudowska-Sawczuk M, Mroczko B. What’s at the moment identified in regards to the function of CXCL10 in SARS-CoV-2 an infection? Int J Mol Sci. 2022;23(7). doi:10.3390/ijms23073673
41. Chen Y, Wang J, Liu C, et al. IP-10 and MCP-1 as biomarkers related to illness severity of COVID-19. Mol Med. 2020;26(1):97. doi:10.1186/s10020-020-00230-x
42. Kalinina O, Golovkin A, Zaikova E, et al. Cytokine storm signature in sufferers with reasonable and extreme COVID-19. Int J Mol Sci. 2022;23(16). doi:10.3390/ijms23168879
43. Heller EA, Liu E, Tager AM, et al. Chemokine CXCL10 promotes atherogenesis by modulating the native steadiness of effector and regulatory T cells. Circulation. 2006;113(19):2301–2312. doi:10.1161/CIRCULATIONAHA.105.605121
44. Koten Ok, Hirohata S, Miyoshi T, et al. Serum interferon-gamma-inducible protein 10 degree was elevated in myocardial infarction sufferers, and negatively correlated with infarct dimension. Clin Biochem. 2008;41(1–2):30–37. doi:10.1016/j.clinbiochem.2007.10.001
45. Wang X, Yue TL, Ohlstein EH, Sung CP, Feuerstein GZ. Interferon-inducible protein-10 includes vascular easy muscle cell migration, proliferation, and inflammatory response. J Biol Chem. 1996;271(39):24286–24293. doi:10.1074/jbc.271.39.24286
46. Taub DD, Lloyd AR, Conlon Ok, et al. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177(6):1809–1814. doi:10.1084/jem.177.6.1809
47. Zabini D, Nagaraj C, Stacher E, et al. Angiostatic elements within the pulmonary endarterectomy materials from persistent thromboembolic pulmonary hypertension sufferers trigger endothelial dysfunction. PLoS One. 2012;7(8):e43793. doi:10.1371/journal.pone.0043793
48. Lupieri A, Smirnova NF, Solinhac R, et al. Easy muscle cells-derived CXCL10 prevents endothelial therapeutic by PI3Kgamma-dependent T cells response. Cardiovasc Res. 2020;116(2):438–449. doi:10.1093/cvr/cvz122






